

Screenshot of two JAABA experiment directories.
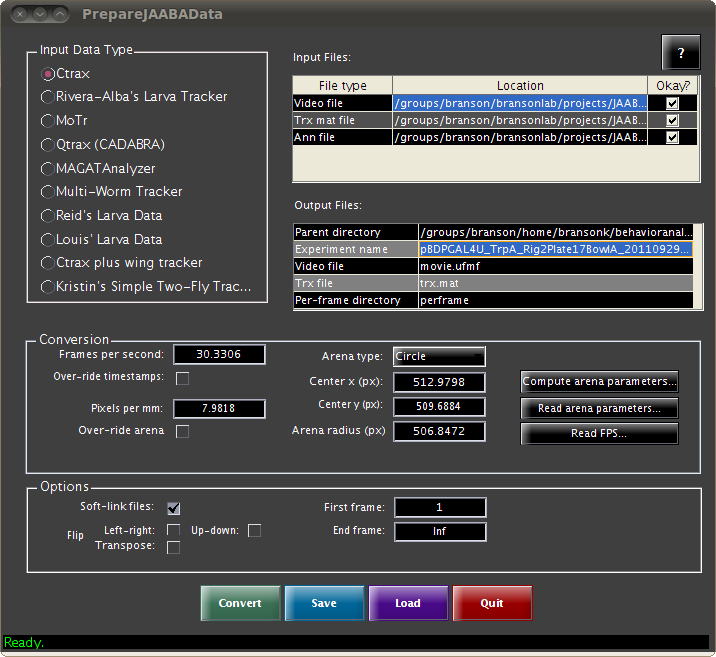
On this page, we describe the format of the data JAABA uses. We provide a program called PrepareJAABAData for converting data output from several trackers to JAABA's format.
The inputs to JAABA are the video and the animals' trajectories in the videos. JAABA terms all data associated with a given video an experiment. All data associated with a single video (the video, the trajectories file, and other data used within JAABA) must be stored in a single, separate directory, called an experiment directory. JAABA uses the directory's name to refer to this experiment. Within this directory, all the files must be named consistently. For example, the filename for the video for different experiments should always be the same (e.g., movie.avi). JAABA can open avi, seq, mov, ufmf, fmf, mmf, and sbfmf video formats. The PrepareJAABAData program can be used to convert the outputs from several existing trackers to the experiment directory structure used by JAABA. Users familiar with MATLAB programming can refer to the tracks file structure below.
The screenshots shows 2 JAABA experiments:
pBDPGAL4U_TrpA_Rig1Plate15BowlB_20110922T145928 and
pBDPGAL4U_TrpA_Rig2Plate14BowlD_20110615T164545. Each contains the following:


With JAABA, we have included a standalone program called PrepareJAABAData which allows users to create experiment directories compatible with JAABA from the outputs of several existing tracking systems.
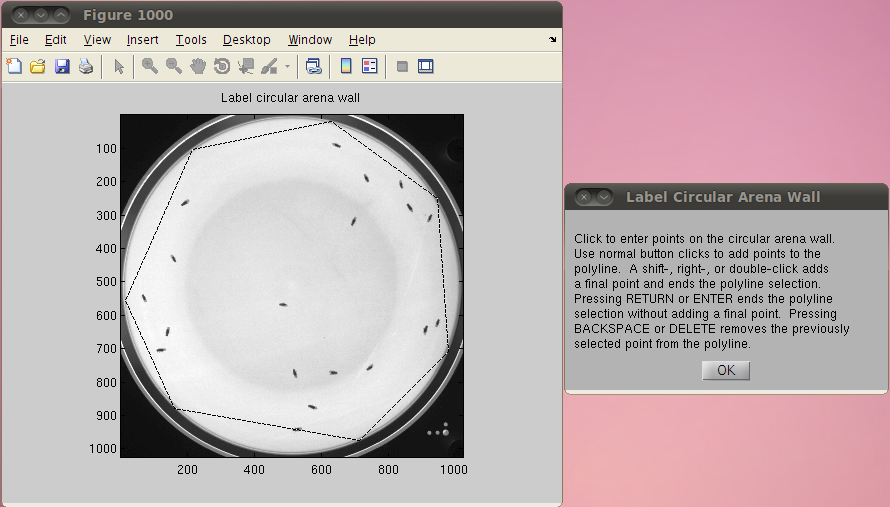
trx. trx is a MATLAB structure array where each element corresponds
to an animal. The details of the structure fields are listed in the table below.
| Structure Field | Description |
|---|---|
x | x-coordinate of the animal in pixels (1 x nframes). |
y | y-coordinate of the animal in pixels (1 x nframes). |
theta | Orientation of the animal (head) (1 x nframes). |
a | 1/4 of the major-axis length in pixels (1 x nframes). |
b | 1/4 of the minor-axis length in pixels (1 x nframes). |
nframes | Number of frames in the trajectory of the current animal (scalar). |
firstframe | First frame of the animal's trajectory (scalar). |
endframe | Last frame of the animal's trajectory (scalar). |
off | Offset for computing index into x, y, etc. Always equal to 1 - firstframe (scalar). |
id | Identity number of the trajectory (scalar). |
x_mm | x-coordinate of the animal in mm (1 x nframes). |
y_mm | y-coordinate of the animal in mm (1 x nframes). |
theta_mm | Orientation of the animal in real coordinates. This is often the same as theta, if no transformation other than translation and scaling is performed between pixels and real coordinates (1 x nframes). |
a_mm | 1/4 of the major-axis length in mm (1 x nframes). |
b_mm | 1/4 of the major-axis length in mm (1 x nframes). |
sex | Sex of the animal. Can be just one value ('M' or 'F' or '?') or a cell array of 'M' and 'F' giving the sex for each frame. The size of the cell array should be nframes. |
dt | Difference in timestamps of the current frame and next frame, in seconds (1 x nframes-1). |
fps | Average frames-per-second (scalar). |
timestamps | Timestamp of each frame (optional), in days (1 x nframes). |
area |
Larvae only. Area of the larva in pixels (1 x nframes). |
xcontour |
Larvae only. x-coordinates of the contour of the larva in pixels (1 x nframes cell array). |
ycontour |
Larvae only. y-coordinates of the contour of the larva in pixels (1 x nframes cell array). |
xspine |
Larvae only. x-coordinates of the spine fit to the larva in pixels (nspinepts=11 x nframes). |
yspine |
Larvae only. y-coordinates of the spine fit to the larva in pixels(nspinepts=11 x nframes). |
area_mm |
Larvae only. Area of the larva in mm2 (1 x nframes). |
xspine_mm |
Larvae only. x-coordinates of the spine fit to the larva in mm (nspinepts=11 x nframes). |
yspine_mm |
Larvae only. y-coordinates of the spine fit to the larva in mm(nspinepts=11 x nframes). |
Below is a screenshot of the MATLAB command window showing the trx variable.
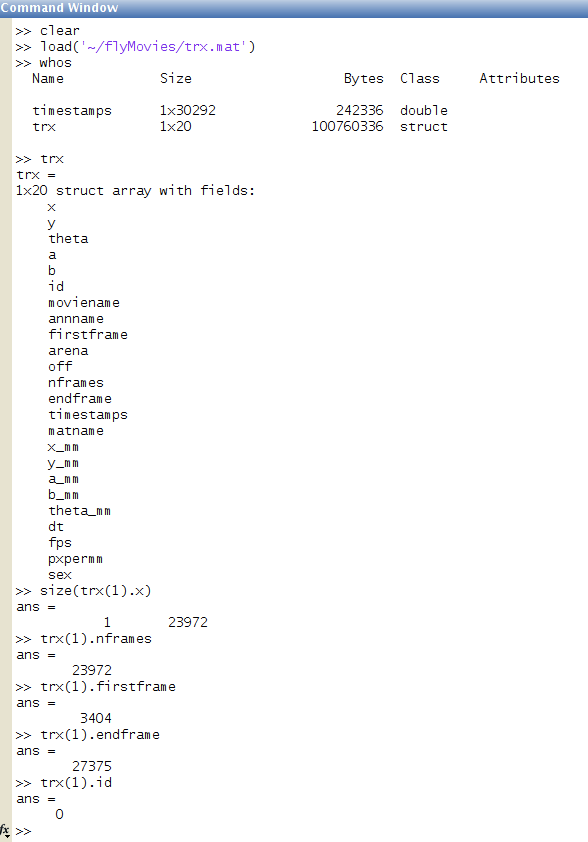
trx variableThe optional variable timestamp in the trxfile gives timestamps for each
frame of the movie, represented as a serial date number (described in
the table above).